Tags#
The tags used in the posts so far are:
catboost (1)#
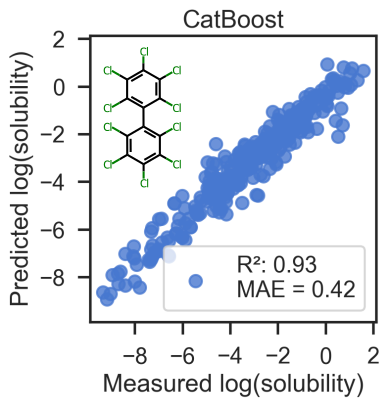
This is an example of using gradient boosting (CatBoost) for predicting solubilities of molecules using molecular descriptors from RDKit.
cheminformatics (2)#

This is an example of using gradient boosting (CatBoost) for predicting solubilities of molecules using molecular descriptors from RDKit.

RDKit can calculate molecular fingerprints and depict the active bits. Here is an example that extends the bit-drawing by RDKit to also show the full molecule.
chemistry (5)#
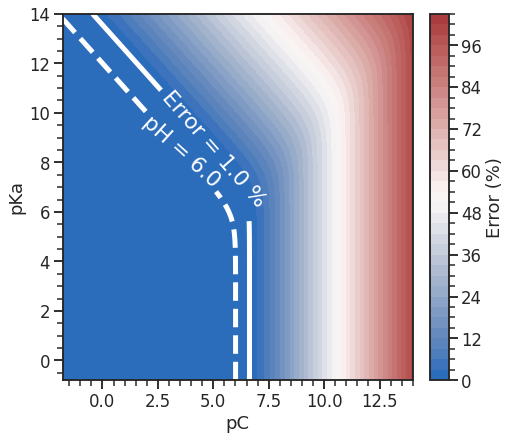
I recently had to go over some details on pH-calculations in weak acids. How can we calculate the pH exactly (with concentrations) and when can we not rely on "standard" solution method we learn in general chemistry? Let us find out.

This is an example of using gradient boosting (CatBoost) for predicting solubilities of molecules using molecular descriptors from RDKit.

This is an example of how one can calculate molecular orbitals (with pyscf) and visualize them.

RDKit can calculate molecular fingerprints and depict the active bits. Here is an example that extends the bit-drawing by RDKit to also show the full molecule.

RDKit and py3Dmol are very useful for working with molecules. Here is a short example of how these libraries can be used to visualize the 3D structure of molecules.
chemometrics (1)#

This is an example of using gradient boosting (CatBoost) for predicting solubilities of molecules using molecular descriptors from RDKit.
descriptors (1)#

This is an example of using gradient boosting (CatBoost) for predicting solubilities of molecules using molecular descriptors from RDKit.
fingerprint (1)#

RDKit can calculate molecular fingerprints and depict the active bits. Here is an example that extends the bit-drawing by RDKit to also show the full molecule.
jupyter (4)#

This is an example of using gradient boosting (CatBoost) for predicting solubilities of molecules using molecular descriptors from RDKit.

This is an example of how one can calculate molecular orbitals (with pyscf) and visualize them.

RDKit can calculate molecular fingerprints and depict the active bits. Here is an example that extends the bit-drawing by RDKit to also show the full molecule.

RDKit and py3Dmol are very useful for working with molecules. Here is a short example of how these libraries can be used to visualize the 3D structure of molecules.
machine-learning (1)#

This is an example of using gradient boosting (CatBoost) for predicting solubilities of molecules using molecular descriptors from RDKit.
molecules (4)#

This is an example of using gradient boosting (CatBoost) for predicting solubilities of molecules using molecular descriptors from RDKit.

This is an example of how one can calculate molecular orbitals (with pyscf) and visualize them.

RDKit can calculate molecular fingerprints and depict the active bits. Here is an example that extends the bit-drawing by RDKit to also show the full molecule.

RDKit and py3Dmol are very useful for working with molecules. Here is a short example of how these libraries can be used to visualize the 3D structure of molecules.
orbitals (1)#

This is an example of how one can calculate molecular orbitals (with pyscf) and visualize them.
py3dmol (1)#

RDKit and py3Dmol are very useful for working with molecules. Here is a short example of how these libraries can be used to visualize the 3D structure of molecules.
python (4)#

This is an example of using gradient boosting (CatBoost) for predicting solubilities of molecules using molecular descriptors from RDKit.

This is an example of how one can calculate molecular orbitals (with pyscf) and visualize them.

RDKit can calculate molecular fingerprints and depict the active bits. Here is an example that extends the bit-drawing by RDKit to also show the full molecule.

RDKit and py3Dmol are very useful for working with molecules. Here is a short example of how these libraries can be used to visualize the 3D structure of molecules.
rdkit (3)#

This is an example of using gradient boosting (CatBoost) for predicting solubilities of molecules using molecular descriptors from RDKit.

RDKit can calculate molecular fingerprints and depict the active bits. Here is an example that extends the bit-drawing by RDKit to also show the full molecule.

RDKit and py3Dmol are very useful for working with molecules. Here is a short example of how these libraries can be used to visualize the 3D structure of molecules.
shap (1)#

This is an example of using gradient boosting (CatBoost) for predicting solubilities of molecules using molecular descriptors from RDKit.